|
|
| Plant Mitosis |
|
Sexual Reproduction Sex is the joining of two genomes from different individuals through the fusion of cells (syngamy) and a later reduction to single genomes in progeny cells by the special cell division, meiosis. Syngamy doubles the number of genomes within the resultant cell (referred to as the zygote). One genome is defined as a single set of chromosomes that includes a complete "library" of genes. A cell with a single genome is referred to as a haploid cell (N) and the resulting zygote with two complete sets of chromosomes (two genomes) is referred to as a diploid cell (2N). Immediately following the beginning of syngamy and before the fusion of the chromosomes from two different nuclei the cell is referred to as N + N. 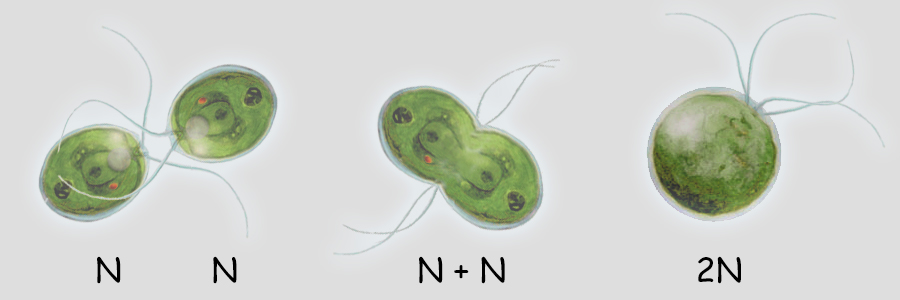 The unicellular green alga, Chlamydomonas (above), typically carries only a single genome of 17 chromosomes when water conditions are good for growth. It is "less expensive" to replicate one set of chromosomes and resources can be directed more to increased cell divisions and increasing population numbers. During times when nutrients for growth become scarce, cells with a single genome join and fuse (syngamy) to become cells with a double genome set of 17 + 17 or 34 chromosomes. The resultant diploid zygote then will encyst entering a resistant state until optimal conditions return. Within this cyst, the zygot will divide by meiosis to produce four haploid cells which will be released to again grow into a population of haploid cells. There is great value to entering the diploid state when conditions worsen as will be discussed later. A simplified illustration of syngamy (below) shows two cells undergoing syngamy each with a single set of 2 chromosomes. The resultant cell (zygote) is diploid (2N) with a double genome set of 2 + 2 or 4 chromosomes. 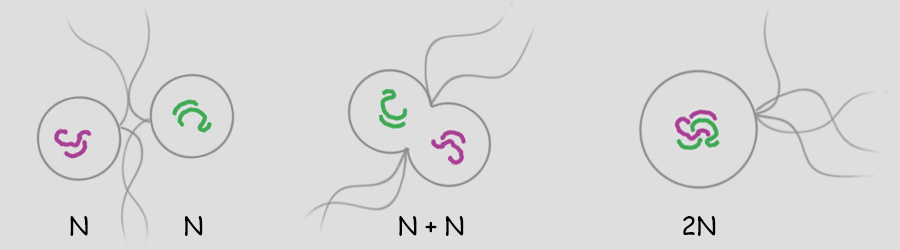 Without meiosis (a special "reduction division" discussed later) repeated syngamy would increase the genome size exponentially from generation to generation. This, of course, is an untenable situation leading to the rupture of the cell. Meiosis is required to return the genome to a single set of chromosomes. Two haploid cells undergo syngamy to become a single diploid cell and diploid cells can undergo meiosis to produce progeny that are haploid. Sexual reproduction requires both syngamy and meiosis. Both haploid cells and diploid cells can divide by mitosis, a cell division that first replicates the genome(s) and then separates the replicated chromosomes into two daughter cells. Mitosis is an ancient type of cell division that the first unicellular eukaryotic cells employed to divide and to reproduce asexually. Most likely, the first eukaryotes were haploid containing a single genome (one set of chromosomes) and used mitosis as a means of asexual reproduction. Multicellular eukaryotes use mitosis to increase numbers of cells, for growth and for wound healing. Terminology Some terminology becomes important at this point in the discussion. A single set of chromosomes contains all of the genes of the nuclear genome. A haploid cell has one genome, a complete set of genes contained within one or more DNA strands. The number of DNA strands or chromosomes is represented by "N". In the illustration above and below, N=2. For the most part, the number of genes and chromosomes and the arrangement of the genes on the chromosomes are the same for all members of a species. In the example above, using green and purple to color-code each haploid set of chromosomes, the two long chromosomes contain the same genes in the same sequence and the two short chromosomes contain the remaining genes and in the same sequence. This does not mean that the chromosomes are identical. A gene may exist in different forms within a population, some members possessing one variant while others possess another variant. For example, in Mendel's experiments with Garden Peas (Pisum sativum), there were flower-color genes located on chromosome 2; however, some pea plants had white flowers and some pea plants had purple flowers. Nearly 50 years later, the peas with white flowers were found to have flower-color genes with a single mutation in which a nucleotide G (guanosine) was replaced with a nucleotide A (adenosine). The wild-type purple gene and the mutant white gene are two forms of the same flower-color gene and are referred to as alleles of the flower-color gene (allo-, different). If a chromosoome 2 with a white allele is lined up with a chromosome 2 with a purple allele, the two alleles will be side-by-side at the same location (locus). So it is with all of the other genes on chromosome 2. Two chromosomes containing the same genes are said to be homologous (homo-, same; -logus, correlation) or homologs of one another. Conversely, in the illustration above and below, a long and a short chromosome do not contain the same genes and are said to be heterologous (hetero-, different; -logus, correlation). 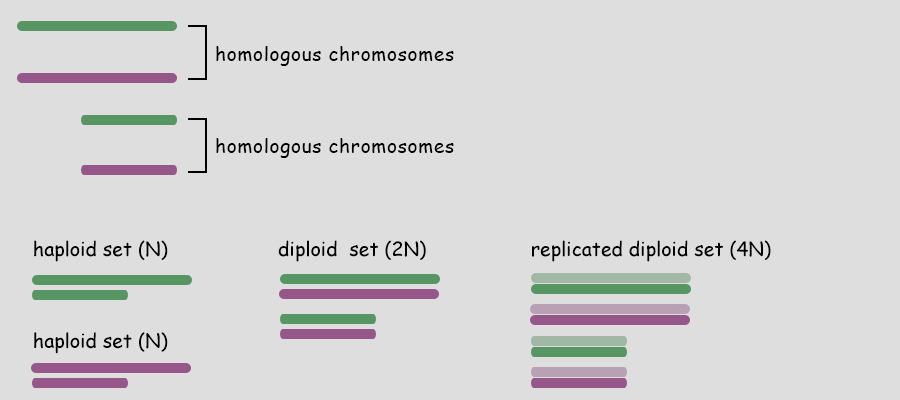 During syngamy, when two haploid cells fuse, they create a cell with two sets of chromosomes, one set from each haploid cell. The resultant cell is referred to as a zygote (zygo-, to join, to yoke) and is now diploid (2N) with two genomes (two sets of chromosomes). In the example above, N + N = 2N. A somewhat confusing terminology is associated with the DNA replication of a diploid cell prior to division. Clearly, in our example, doubling the number of chromosomes from four to eight makes the cell tetraploid (4N); however, recognizing that replicated chromosomes are identical and still joined together, the chromosomes of each pair are referred to not as chromosomes, but as sister chromatids and the cell is still considered diploid. But that is just terminology; the cell really is tetraploid.
As mentioned, a single genome is one entire set of nuclear genes, but they are distributed along the lengths of different numbers of chromosomes. For example, the human genome comprises about 19,000 genes organized within a haploid set of 23 chromosomes. On the other hand, the Coyote has about the same number of genes, but they are organized within a haploid set of 39 chromosomes. Corn (Maize) has abnout 32,000 genes organized into a haploid set of 10 chromosomes. Alfalfa has about 40,000 genes organized into a haploid set of 8 chromosomes. The number of chromosomes in a haploid set can vary from one, as in many bacteria and some ants, to several hundred, as in some ferns like Ophioglossum (N=634), the Mulberry (N=154) and the diminutive blue butterfly in the genus Agrodiaetus (N=134). Mitosis of a Haploid Cell: Simplified The essential aspects of mitosis are that (1) chromosomes (strands of DNA) replicate prior to mitotic cell division and condense during prophase such that they become visible as two strands bound together at the centromere. This is illustrated as a pale and dark member of the replicated pair. The two chromosomes, while attached to one another, are referred to as sister chromatids. (2) During metaphase the replicated chromosomes line up in the center of the cell and microtubules (aka, spindle fibers) bind to the centromeres. (3) During anaphase the chromatids as separated, and the microtubules pull them in opposite directions to the poles of the cell. (4) During telophase the cell divides, isolating the separated sister chromatids into their own cells. The sister chromatids are now referred to as chromosomes. In the illustration below, the cell began with one genome comprising a long and short chromosome and divided to become two cells each with one genome, a long and short chromosome. The number of chromosomes comprising a single genome varies widely from species to species. A genome of only two chromosomes is shown only for simplicity. 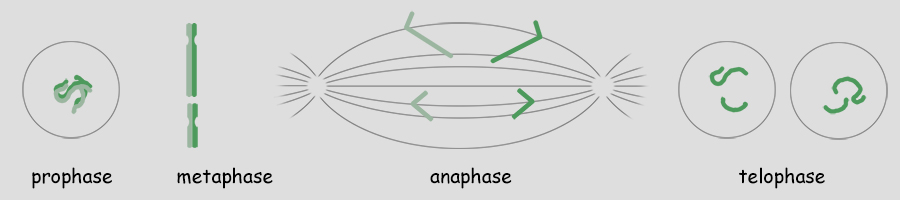 As occasionally happens by mistake, the separation of sister chromatids does not complete in a timely manner and both sister chromatids are pulled together to one pole of the cell. This could occur for one or both pairs and the pairs could be pulled in the same direction or in opposite directions. The illustration below shows both pairs of conjoined sister chromatids being pulled in the same direction, resulting in one cell without chromosomes (which will die) and one with a doubled genome set . . . with both sets being identical to one another. This need not be a lethal condition. New species have evolved due to this error. The failure of sister chromatids to separate is referred to as nondisjunction and results in a cell with multiple genomes, a condition referred to as polyploidy (having three or more genome sets) or as aneuploidy (having an abnormal set of chromosomes). Some new species have had their origins in just this fashion; it is common among plants. And, having a "back-up" set of chromosomes protects against mutations in one or the other genome that would be harmful. Due to the regular rate of mutations, the two sets of chromosomes will slowly become different from one another, providing a new "palette" of characteristics. In fact, polyploidy has shaped the evolution of a number of eukaryotes. Oilseed rape (a member of the mustard family), cotton, wheat, goldfish, the grey treefrog and many other species are polyploid due to nondisjunction or a variety of other mechanisms. 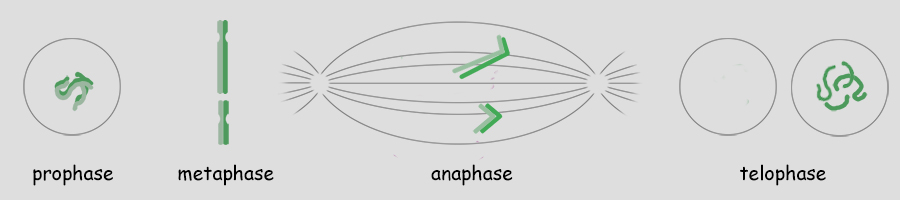 Above. Nondisjunction. (I know, I know, why not simply call it non-separation.) If both pairs of sister chromatids are pulled to the same pole, one cell will certainly die because it will have no chromosomes. The other may well survive with two sets of chromosomes. If the two pairs of chromosomes do not separate and are pulled to opposite poles, both cells will die because neither will have a complete genome. Some organisms, particularly fungi, have mechanisms for "throwing out" extra chromosomes to revert to a single genome. Aneuploidy, as the result of nondisjunction, is the most common chromosome abnormality in humans and is the leading cause of miscarriages and congenital birth defects. I only mention this here to emphasize that nondisjunction, although somewhat uncommon, is not rare, the effects occurring in the sperm of about 1 in 500 men and with increasing frequency with age in the eggs of women. Mitosis of a Diploid Cell The stages and processes of mitosis are the same whether the cell is haploid with one genome set of chromosomes or is diploid with two genome sets of chromosomes. Chromosomes enter mitosis replicated and condense during prophase, align individually on the central plane bisecting the cell during metaphase, and, once the sister chromatids are freed, they are drawn to opposite poles during anaphase. Photomicrographs from the Lily (below) show the stages of mitosis and the appearance of the chromosomes at each stage. The Lily is diploid with 24 chromosomes in the cells of the leaves, roots and stems, two sets of 12 chromosomes. N = 12; 2N = 24. 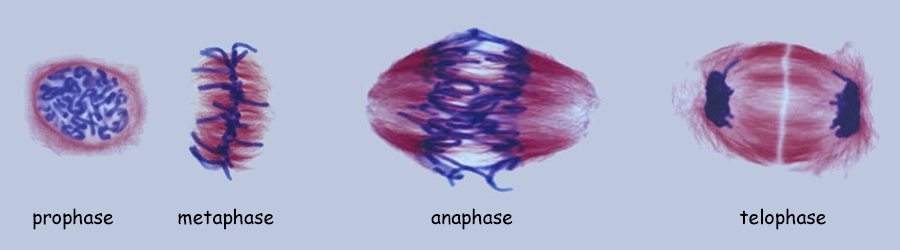 Beginning with a hypothetical cell that has two genomes of two sets of two chromosomes each, identified graphically by two colors, green and purple, the basic principles of mitosis can be identified more simply than by following a cell like that of the Lily with 24 chromosomes. Prior to mitotic cell division the chromosomes replicate, the same as occurred in the previous example for mitosis of a haploid cell with one set of chromosomes. 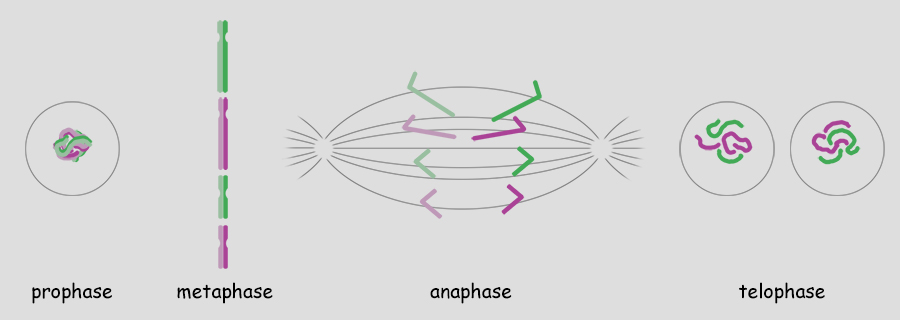 At metaphase, all replicated chromosomes line up individually on an equatorial plane that bisects the cell. Not shown in the illustration above, microtubules from opposite poles bind to the centromeres and bring them to the central, metaphase position. Once the centromere region of the chromosome dissociates, the sister chromatids are now free to be pulled to opposite poles during anaphase. The alignment and separation of sister chromatids during mitosis is referred to as karyokinesis (karyo-, kernal or nucleus; -kinesis, movement). With the now single chromosomes at opposite poles, what remains is the division of the cytoplasm into two cells during telophase. The division of the cell following karyokinesis is referred to as cytokinesis. The result of the full mitotic process is two cells with the same number of chromosomes as the parent cell, although unreplicated. Before the next mitotic division, the chromosomes once again will replicate. The stage or phase wherein a cell performs its normal functions (is not dividing) and may be preparing for a future cell division is referred to as interphase. Stages of Mitosis Mitosis is a continuous process of chromosome movement flowing uniformly through the alignment and separation of chromosomes (mitosis) and the division of the cell into two (cytokinesis). The stages of mitosis are hallmark scenes in this "movie" that have been clipped out to create a summary storyboard. The main stages are interphase, prophase, metaphase, anaphase and telophase (often written on student desks as IPMAT in order to remember the sequence during exams). Mitosis is the process by which the chromosome sets are organized and isolated to oppposite ends of the cell; the process is often referred to as karyokinesis (nuclear action). Once sparated from one another, plant chromosomes are finally isolated in daughter cells by the formation of a wall that divides the cells. This final, actual division of the cell, is referred to as cytokinesis (cell action). Of course, one could more finely divide the "script" of mitosis and arbitrarily create more stages and often this is done by referring, for example, to early and late propahse or to prometaphase, etc.
During the S subphase of interphase, the DNA of chromosomes is replicated and the two DNA strands produced are held together in a region referred to as the centromere. From the S phase through mitosis, the cell will have two sets of DNA until the two sets are separated at telophase and partitioned between the two daughter cells during cytokinesis. Following S subphase and while held together, the fidelity of replication is checked and if errors have occurred repairs will be attempted before mitosis can continue. If the errors are significant and cannot be repaired, the cells will die. 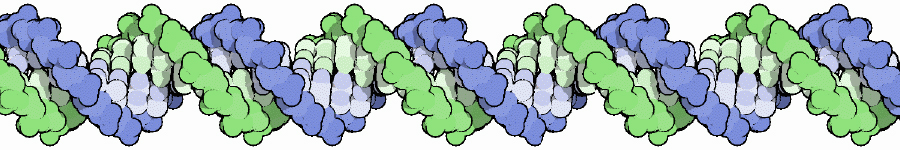 Above. A molecule of DNA is actually two molecules entwined with one another in the famous "double helix". At the core of each chromosome is a double stranded DNA molecule. If the chromosome number of a cell is 10, then the number of DNA molecules is 10. The problem experienced by a dividing cell is that the DNA molecules are extremely long . . . each can be as long as 3 feet or more . . . and with multiple strands of DNA (chromosomes) in the nucleus of cells, separation of the strands would be difficult. It once was thought that chromosomes were a tangled mass dispersed throughout the interphase nucleus, like a bowl of spaghetti. Now it is known that, although extended, the DNA molecules are organized within "chromosome territories" within the nucleus and only moderately overlap with nearest neighbors. Still, the DNA strands must be more compact in order for the daughter chromsomes (chromatids) to be organized and separated. Chromosome Condensation In order for DNA replication to occur, the DNA strands must be extended and in order to separate the chromosomes following replication, they must be condensed into manageable lengths. Replication of DNA occurs in the S subphase of interphase and packaging and condensation of the DNA into chromosomes occurs during prophase. Following the S subphase and the successful repair of any errors, the DNA strands begin to condense and become visible as individual chromosomes. Prior to this condentation, the DNA strands are extended and beyond the resolution of the light microscope. 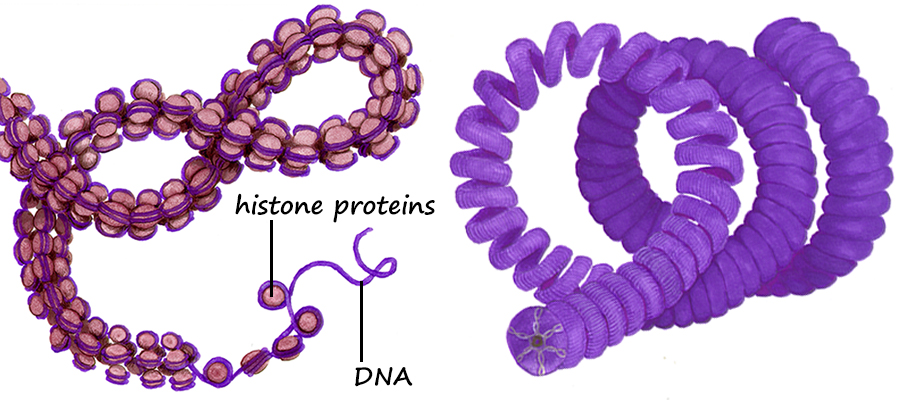 Above. Following the replication of DNA in the S subphase of interphase and during the subsequent prophase, histone proteins (represented by tan spheres) are modified and begin to bind DNA in groups of eight histone proteins. The DNA wraps the the eight histones twice to form DNA-protein structures referred to as nucleosomes. Extending from the histones are the free ends of the protein chains. Free DNA between nucleosomes is termed "linker DNA". 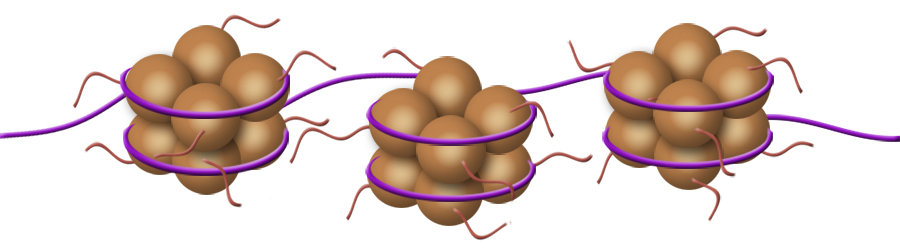 Above. Condensation continues with modifications of specific amino acids of the free protein ends of the histones. These modifications allow two condensing proteins (condensin I and condensin II) to bind and to begin joining the nucleosomes into organized loops around a central axis. Continued condensation results in the much shorter protein-DNA complexes called chromosomes. 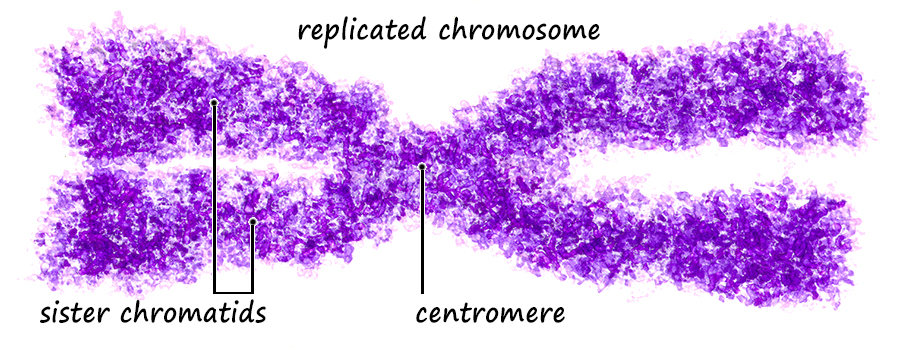 The two double-stranded DNA molecules that are the result of DNA synthesis (replication) remain coupled together in a region referred to as the centromere. Centromeres, Kinetochores and Microtubules The region where sister chromatids are joined is referred to as the centromere which means "central segment"; however, centromeres can be at various locations along the chromosome from the middle to the end. With few exceptions, each chromosome has a single centromere location and the centromere's location helps to differentiate the chromosome from other chromosomes that have centromeres at different locations. The centromere is a specialized region of DNA to which centromere binding proteins (CENP proteins) attach. The nucleotide sequences of the centromere are distinctive and different from other regions. There are more than twenty proteins described that bind to centromere DNA or to other proteins that bind to special base pair sequences of DNA in this region. In the region of the centromere, a variant histone protein (CENP-A) creates novel nucleosomes that begin the condensation of centromere DNA. Another sequence of about 170 base pairs contains within it a 17-base-pair sequence, referred to as the "centromere protein B box" (CENP-B box). A single centromere may contain from hundreds to thousands of these regions each with a CENP-B box. The CENP-B boxes bind centromere protein B (CENP-B) which is a protein comprising two identical strands, each of which can bind to a CENP-B box which allows the cross-linking of the two chromatids in the region of the centromere. In addition, another CENP, cohesin, 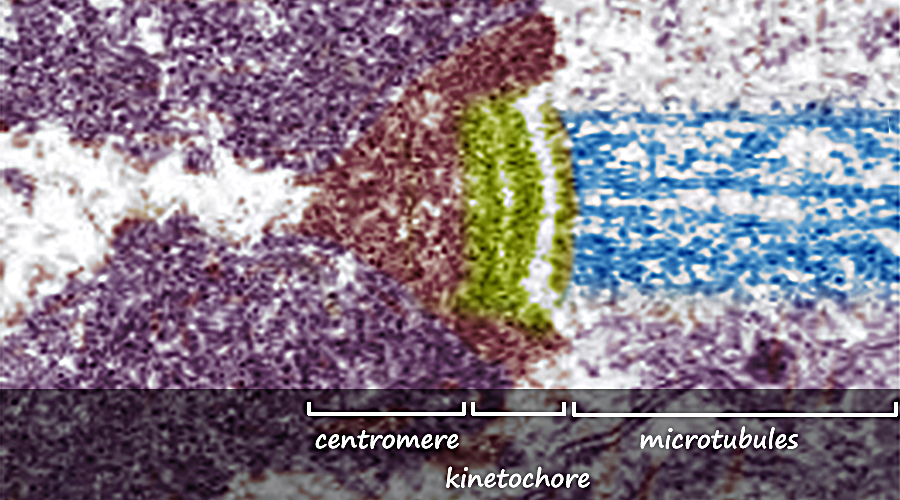 Above. The centromere and kinetochore regions of an anaphase chromosome that is being pulled to the right. Additional proteins bind to the CENPs that are bound to the centromere DNA. Many of these additional proteins comprise a complex structure referred to as the kinetochore. The kinetochore is a structure associated with the centromere to which microtubules can bind. Microtubules are the spindle fibers that will separate the sister chromatids during anaphase. Summary of Stages of Mitosis |

| Early Prophase As the interphase cell enters prophase the protein-DNA complexes within the nuclear envelope become visible with staining. The chromosomes begin to be seen as having been replicated with two chromosomes bound together at the centomere. Outside the nucleus, tubulin proteins begin to form microtubules that will become the spindle fibers that will separate the daughter chromosomes. |

| Prometaphase The disintegration of the nuclear envelope frees the chromosomes into the cytosol and signals the end of prophase. Microtubules (spindle fibers) from opposite ends (poles) of the cell begin attaching to centromeres and attempt to pull them in opposite directions. |

| Early Metaphase The "tug-o-war" between the spindle fibers causes the replicated chromosomes to align on a central plane (the equatorial plane) at the center of the cell. |

| Metaphase Metaphase is the stage during which the chromosomes are aligned on the equatorial plane by their centromeres |

| Early Anaphase Cohesins and CENP proteins dissociate allowing the chromatids to become separated. As the daughter chromosomes are separated at the centromeres, they begin being pulled to opposite ends of the cell. |

| Anaphase The daughter chromosomes become separated and drawn toward opposite poles. This stage assures that each new cell will receive one member of each replicated pair. That is, each daughter cell will receive a complete set of chromosomes. |

| Early Telophase Each chromatid begins to de-condense and to become consolidated within the regions that will be become the two new nuclei. |
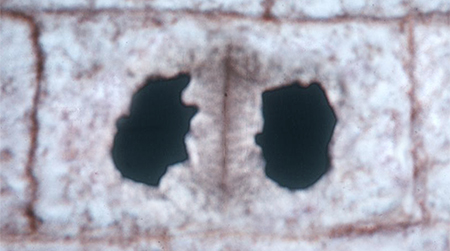
| Telophase Vesicles containing cell wall materials begin to align along the equatorial plane. As they fuse, the cell wall material forms a central plate that will become the cell wall between the new cells. |
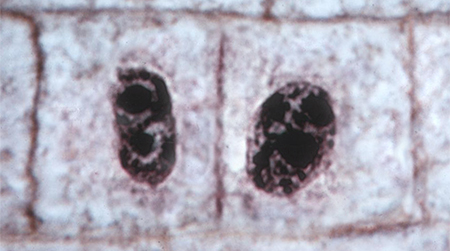
| Cytokinesis The cell wall continues to grow and upon completion two new cells have formed. The chromatids finally are fully extended within their chromosome territories and the nuclear envelopes have formed around the chromosomes. |
| The following was created by dissolving the sequence of images above into a video. |
| |